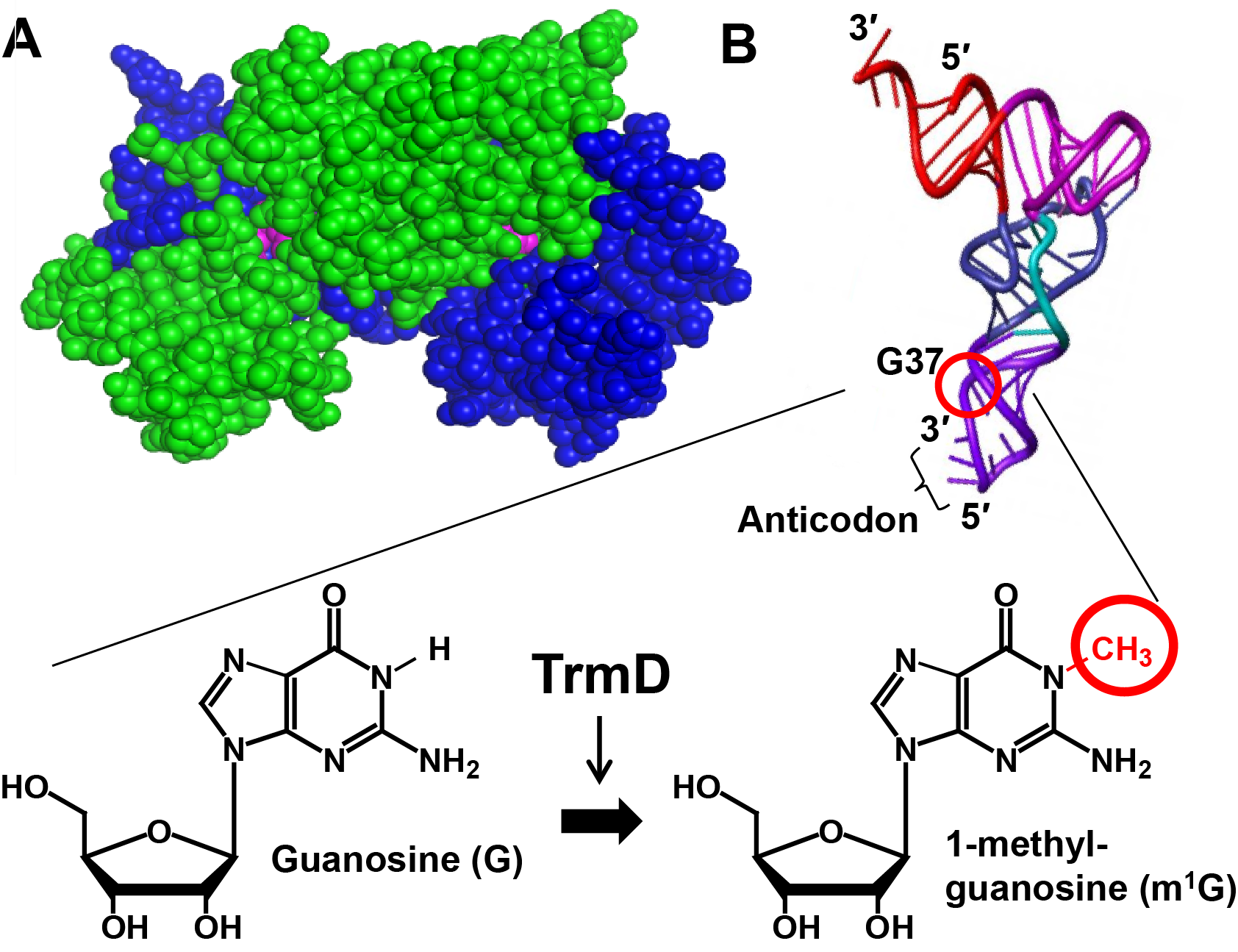
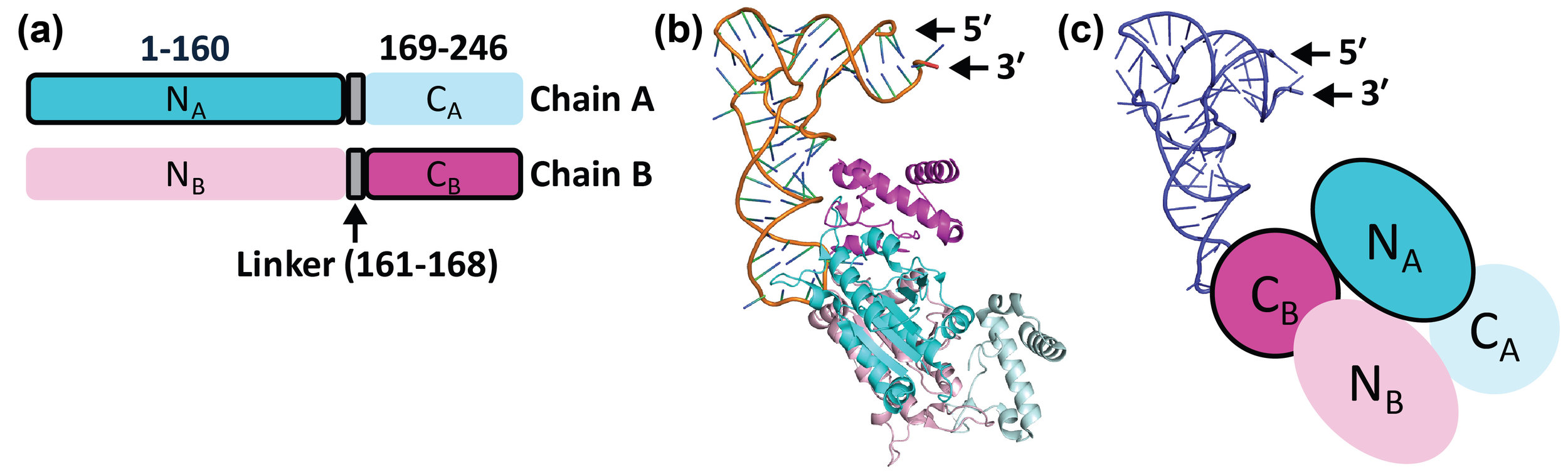
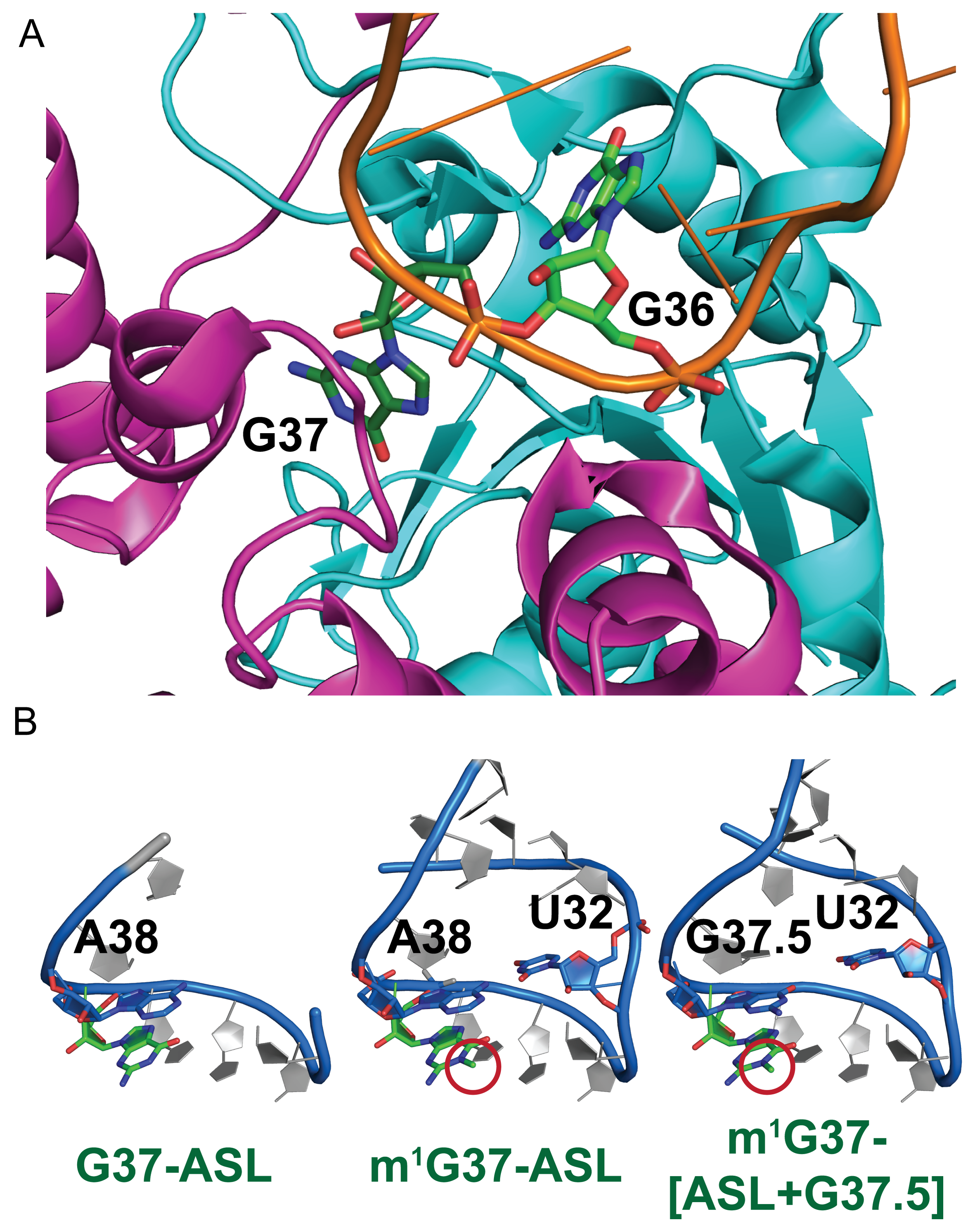
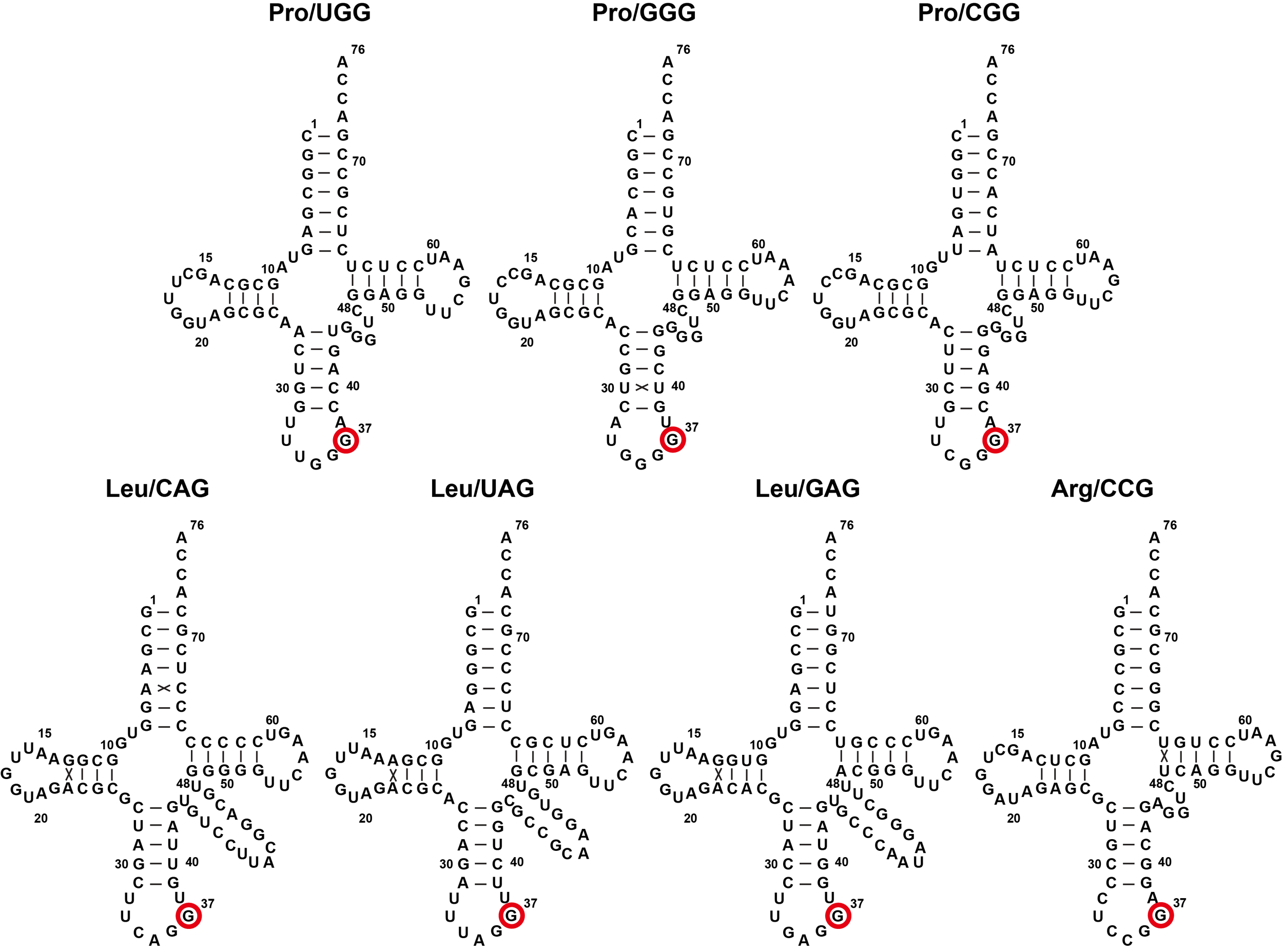
The Hou Laboratory is no longer at this location. Please visit this site for the most current information.
We are interested in the specificity of translation of the genetic code, focusing on the questions of how tRNAs are synthesized, matured, modified, aminoacylated, and function on the ribosome.
The tRNA molecules are essential for the specificity of decoding, which is the key determinant in the speed and quality of cell growth. Elevated levels of tRNAs can lead to cancer, while deficiency in tRNAs can lead to cell toxicity.
Our research provides the basis to gain biochemical, structural, and bioinformatic insights into tRNAs in evolution. These insights are important for understanding the origins of the genetic code and for developing new strategies for drug targeting against diseases arising from errors of tRNA functions.
We have developed a variety of methods, including biochemical, structural, kinetic, and genetic approaches. We focus on one representative enzyme in each case and build a framework of information by examining the enzyme in the larger biological context. Because tRNAs are ancient and enzymes that interact and recognize tRNAs are also ancient, we have a large database to search for related and homologous enzymes in evolution. The tRNA-interacting network is broad and include enzymes and proteins that are in pathways unrelated to protein synthesis. Some of our work published in various journals is mentioned in our publications page.